The ODAP is guided and overseen by the ODAP Partnership, a collaboration among nationwide research studies, public health agencies, the NHS and clinicians and scientists.

What it is
- The purpose of the outbreak data analysis platform is to provide an accessible, usable data resource to enable research relevant to COVID-19 and future outbreaks. This will accelerate scientific understanding of new outbreaks for the benefit of patients and the protection of the public.
- It will create a UK-wide capability by curating and linking outbreak relevant data from clinical records, research studies and audit data. It brings together key initiatives and leadership across the UK including ISARIC, COG-UK, MRC CLIMB and GenOMICC.
- The platform combines a national Trusted Research Environment (TRE) infrastructure collocated with >£100M of world-class computational and data science capacity including the UK National Supercomputer, with a UK-wide governance framework.
What it isn’t
- It is not a replacement for public health activities. The platform will work closely with public health agencies across the 4 nations and with UK HSA, providing research insights and data feeds where useful to augment surveillance capacity, and a tried-and-tested route to engage additional analytic capacity and expertise from the academic sector.
- It is not a viral sequence analysis platform. The MRC CLIMB resource (a partner in this programme) provides excellent resources and infrastructure for analysis, presentation and sharing of viral sequences.
- It is not a replacement for existing TREs. The analysis platform is a focused capability of the UKSA-accredited TRE hosted by Public Health Scotland, and will provide curated data feeds to TREs across the UK to facilitate and supplement data held elsewhere as part of a UK federated network.
Design of the platform
Data held
The platform already contains a unique aggregation of UK sovereign data assets, including the complete data resources of the ISARIC4C/CO-CIN, GenOMICC, PHOSP and UK-CIC studies, together with viral sequence data from COG-UK, and linkage to NHS clinical records and structured clinical audit data. This creates a unique opportunity to combine clinical, biological, genomics and virology research in as secure, openly-accessible framework. Manual curation of these linked datasets, in a single platform, is a key step to maximise data quality and usability.
Compute power
The platform is located at the UK National Supercomputer, and uses a range of capabilities from Private Cloud TRE infrastructure to large shared memory systems and massively parallel processing power. There is currently 2.5Pb of storage in use, expandable to many Petabytes as required. GPU servers are in place for massively-parallel applications such as genomics and image analysis
An API structure has been built for real-time data sharing with other trusted research environments across the UK, and to facilitate efficient data pipelines to supply surveillance feeds to public health.
Structure
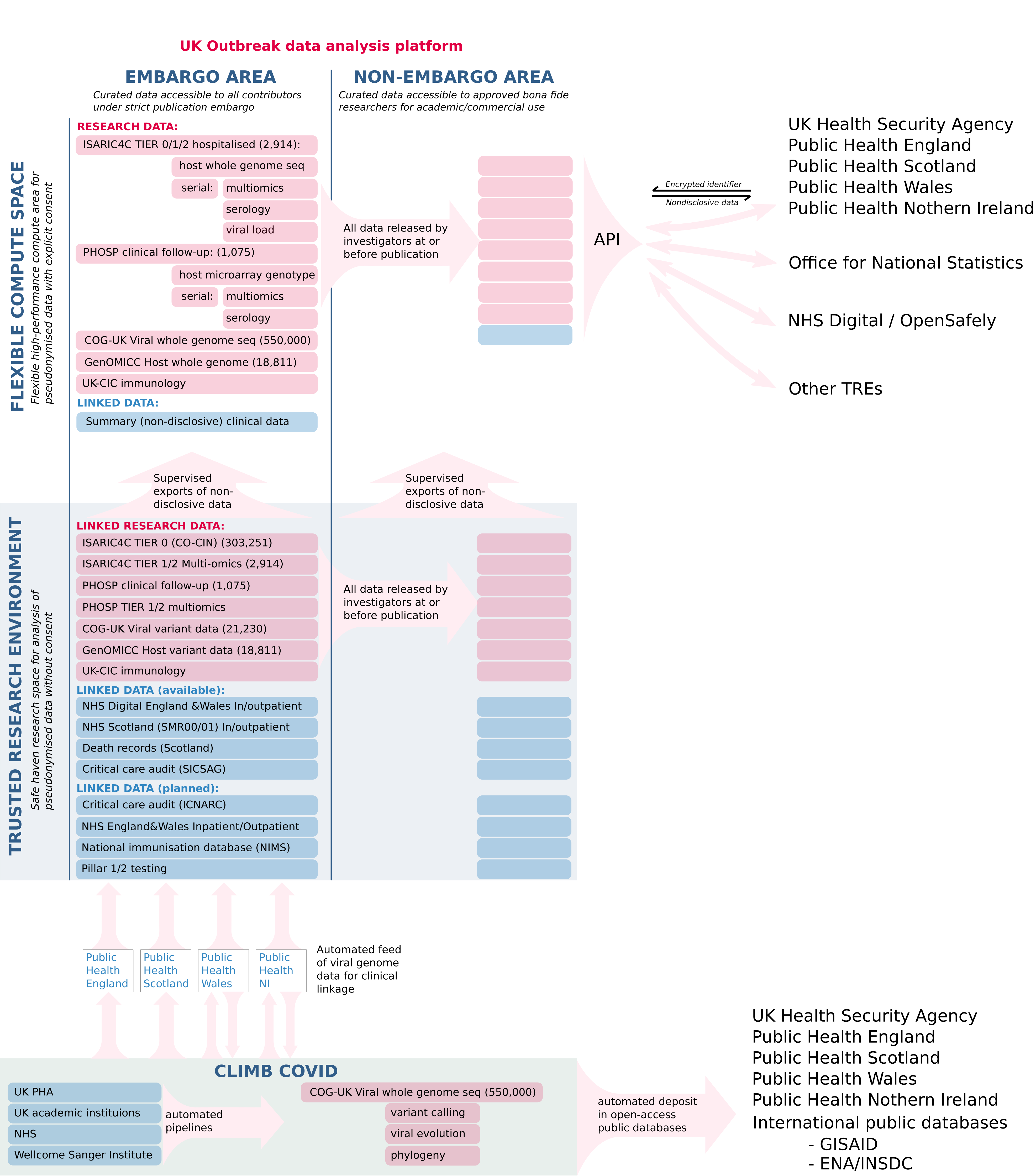
There are two routes of access to the analysis platform (Figure 1):
- NHS Trusted Research Environment (Safe Haven) for access to personal clinical data and data collected without explicit consent.
- Rapid-access flexible compute for access to non-disclosive research data collected with explicit consent.
Within both of these environments there are two levels of access, governed by the data contributors:
- Publishable “open access” data which any user can use and report as they wish, according to data protection and privacy rules;
- Embargoed active research data, shared by academic investigators and available for linked analysis but not for publication without agreement from all contributors.
This design is intended to demonstrate trust in order to encourage immediate contributions of research data from academic collaborators. It makes data available immediately for discovery, whilst protecting the rights of data creators and contributors.
Research outputs
The proposal builds upon an established track record of impact. By generating, integrating and analysing clinical, biological, genetic and virological data on patients with Covid-19 in UK hospitals, the outbreak analysis platform has facilitated research that:
- provided essential weekly updates to SAGE that guide the public health response isaric4c.net/reports/,
- identified host genetic mechanisms of disease,
- quantified the role of age, comorbid illness and obesity in disease severity,
- created the global standard ISARIC4C score for prognostication isaric4c.net/risk,
- determined the impact of long Covid following hospitalisation,
- identified the substantial effect of transmission of Covid-19 within hospitals SPI-M/SAGE report,
- provided key evidence underlying the choice of therapeutic agents for clinical trials,
- provided real world data on vaccine effectiveness and failure (SAGE 87 Egan et al, Egan et al.)
- observed data supporting identification of high risk groups for vaccination (highlighted in No10 briefing)
- described the complications of Covid-19 in hospitalised patients.
Current data content
This platform now serves as a hub for a coordinated UK national research response to COVID-19. Data are included from:
- ISARIC4C tier 0: (unconsented) prospective clinical data from 298,836 cases
- ISARIC4C tiers 1 and 2: serial multiomic assays from research samples of blood, respiratory secretions, urine, and stool from 2,505 cases
- COG-UK: (unconsented) summary variant data from COG-UK viral sequencing study is already included for matched patients
- GenOMICC study complete data: microarray/whole genome sequence data from 18,158 cases
- PHOSP complete data: follow-up clinical and biological data generated by the Post-Hospitalisation for COVID-19 follow-up study (1,075 cases)
- UK-CIC: deep immunological phenotyping data from across the UK Coronavirus Immunology Consortium, using ISARIC4C samples and local collections.
Research data within the analysis platform is linked to:
- NHS Scotland primary, secondary care and death records
- NHS Digital/NHS England health records data
- ICNARC and SICSAG critical care audit databases
- NIMS National Immunisation Dataset
- Pillar 1 Covid-19 testing
- Pillar 2 Covid-19 testing
